ProBioPred Workflow:
Schema:
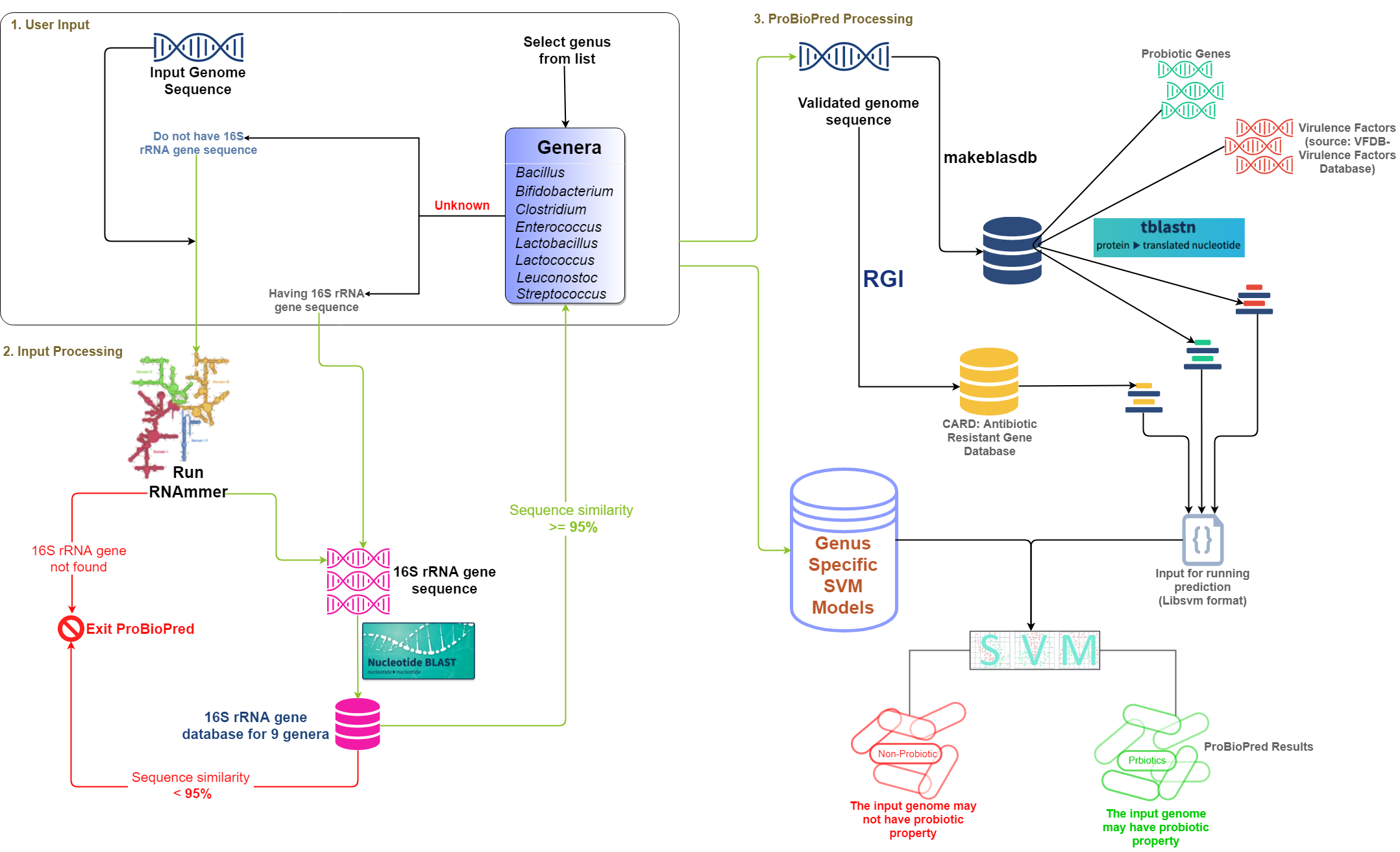
ProBioPred Method:
| Gene name / Locus tag | Functional Category | Role / Function | ProBioPred Catagory |
|---|---|---|---|
| dltA | Cell Envelope | d-Alanylation of LTA | Resistance Acid, Biofilm |
| gadC | Active removal of stressors | Glutamate/ gamma aminobutyrate antiporter | Resistance Acid |
| LBA0996 | Active removal of stressors | Ornithine decarboxylase | Resistance Acid |
| LBA1272 | Cell Envelope | d-Alanylation of LTA | Resistance Acid |
| LBA1524 | 2 CRS Regulators | Histidine protein kinase | Resistance Acid |
| rrp1 | 2 CRS Regulators | Response regulator | Resistance Acid |
| clpL | Protection and repair DNA and proteins | Clp ATPase (chaperone) | Resistance Acid, Resistance Bile |
| LBA0995 | Active removal of stressors | Amino acid permease | Resistance Acid, Resistance Bile |
| LBA0867 | Active removal of stressors | Transcriptional regulator | Resistance Acid, Resistance Bile |
| cdpA | Cell surface Proteins | Cell wall-modifying enzyme involved in cell division and separation | Osmotic, Resistance Bile |
| clpE | Protection and repair DNA and proteins | Clp ATPase (chaperone) | Resistance Bile |
| LBA1427 | Active removal of stressors | Putative oxidoreductase | Resistance Bile |
| LBA1428 | Active removal of stressors | MDR protein (major facilitator family) | Resistance Bile |
| LBA1429 | Active removal of stressors | MDR protein (major facilitator family) | Resistance Bile |
| LBA1430 | 2 CRS Regulators | Histidine protein kinase | Resistance Bile |
| LBA1431 | 2 CRS Regulators | Response regulator | Resistance Bile |
| LBA1432 | Protein with similar to RelA | Response regulator | Resistance Bile |
| dps | Protection and repair DNA and proteins | DNA protection during starvation and other stresses | Resistance Bile |
| slp | Cell Envelope | S-layer protein | Osmotic, Resistance Bile |
| slpA | Cell Envelope | S-layer protein | Osmotic, Resistance Bile, Adherance |
| lspA | Cell surface Proteins | Large surface protein, Putative mucus-binding | Adherance |
| LBA1663-4 | Cell surface Proteins | Putative mucus-binding | Adherance |
| fbpA | Cell surface Proteins | Putative fibronectin-binding | Adherance |
| mub | Cell surface Proteins | Putative mucus-binding | Adherance |
| srtA | Cell surface Proteins | Sortase | Adherance, Competitive |
| msa | Cell surface Proteins | Mannose-specific adhesin | Competitive |
| prtP | Protein metabolism | Cell wall-bound proteinase | Competitive |
| copA | Active removal of stressors | Copper-transporting ATPase | Competitive |
| gtfA | Carbohydrate metabolism, Cell Envelope | Glucosyltransferase | Biofilm, Competitive |
| inu | Cell Envelope, EPS | Inulosucrase | Biofilm, Competitive |
| lp_1403 | 2 CRS Regulators | Histidine protein kinase | Competitive |
| pts14C | Carbohydrate metabolism | Cellobiose PTS, EIIC | Competitive |
| xylA | Carbohydrate metabolism | Xylose isomerase | Competitive |
| met | Protein metabolism | Independent methionine synthase | Competitive |
| clpC | Protection and repair DNA and proteins | Clp ATPase (chaperone) | Persistance |
| Lj1021 | EPS | Entire EPS cluster | Persistance |
| Lj1654 | Carbohydrate metabolism | PTS sugar transporter | Persistance |
| Lj1656 | Carbohydrate metabolism | PTS sugar transporter | Persistance |
| lp_2940 | Carbohydrate metabolism | Sortase-dependent cell wall protein | Persistance |
| luxS | Protection and repair DNA and proteins | Activated methyl cycle | Persistance, Anti-bacterial |
| msrB | Protection and repair DNA and proteins | Methionine sulfoxide reductase | Persistance |
| lsp | Protection and repair DNA and proteins | Putative mucus-binding | Adherance |
| bsh1 | Active removal of stressors | Bile salt hydrolase | Hydrolyze bile-salt |
| bshA | Active removal of stressors | Bile salt hydrolases | Hydrolyze bile-salt |
| bshB | Active removal of stressors | Bile salt hydrolase | Hydrolyze bile-salt |
| LJ0056 | Active removal of stressors | Bile salt hydrolase | Hydrolyze bile-salt |
| dltD | Cell Envelope | d-Alanylation of LTA | Biofilm, Immune-modulation, Simulated Gastric juice |
| wzb | EPS | EPS biosynthesis,phosphotyrosine protein phosphatase | Biofilm |
| iamA | EPS | Response regulator of a 2CRS involved in regulation of EPSs and membrane proteins | Biofilm |
| bfrA | Carbohydrate metabolism | Intracellular fructosidase (FOS metabolism) | Growth |
| fosE | Carbohydrate metabolism | Extracellular fructosidase | Growth |
| treC | Carbohydrate metabolism | Trehalase, osmoprotection | Growth |
| msmE | Carbohydrate metabolism | ABC transporter substrate binding protein | Growth |
| Lr1584 | Active removal of stressors | MDR protein (major facilitator family) | Adaption |
| Lr1265 | Active removal of stressors | Multidrug resistance protein (ABC transporter family) | Adaption |
| labT | Antipathogenic effects | Putative ABC exporter for lactacin B | Anti-bacterial |
| abpT | Antipathogenic effects | Putative ABC exporter for bacteriocin Abp119 | Anti-bacterial |
| cps1A-J | Immunomodulation | capsular polysaccharide | Immune-modulation |
| dltB | Immunomodulation | d-Alanylation of LTA | Immune-modulation |
NOTE: The antibiotic resistance genes (ARGs) used in ProBioPred was taken from Comprehensive Antibiotic Research Database (CARD v2.0.0) database and the Resistance Gene Identifier Software (RGI v4.0.2) utility was used with default parameters to predict the ARGs for input genome. We have used VFDB database for virulence factors.
ProBioPred scoring scheme:
Score for each category of probiotic genes is derived from percent identity of each hit and number of hits found in category (c) as per equation below.
$$Score_{(c)} = {\sum_{i=1}^n \bigl(P_{(i)}) + n}$$
where,
Pi =
Percent identity of ith hit in category (c)
n = Number of
total hits found in category (c)
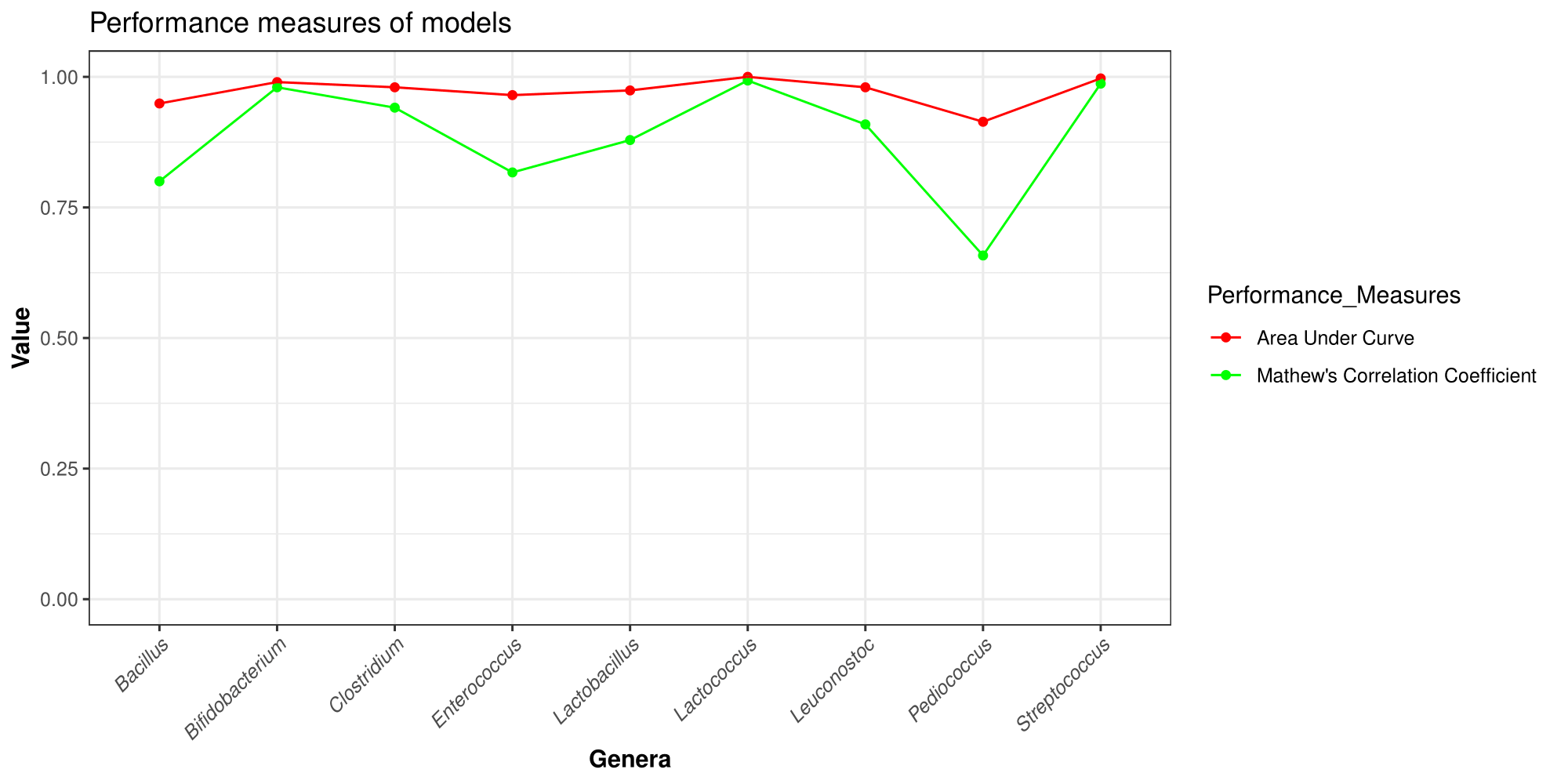
ProBioPred performance:
| #Sr.No. | Genera | Accuracy | Sensitivity | Specificity | Precision | Recall |
|---|---|---|---|---|---|---|
| 1 | Bacillus | 90.08 | 0.914 | 0.891 | 0.862 | 0.914 |
| 2 | Bifidobacterium | 99.23 | 0.99 | 0.995 | 0.995 | 0.99 |
| 3 | Clostridium | 97.04 | 0.955 | 0.984 | 0.98 | 0.955 |
| 4 | Enterococcus | 90.83 | 0.916 | 0.901 | 0.895 | 0.916 |
| 5 | Lactobacillus | 93.95 | 0.938 | 0.941 | 0.939 | 0.938 |
| 6 | Lactococcus | 99.67 | 0.999 | 0.995 | 0.995 | 0.999 |
| 7 | Leuconostoc | 95.43 | 0.944 | 0.965 | 0.965 | 0.944 |
| 8 | Pediococcus | 82.78 | 0.798 | 0.873 | 0.904 | 0.798 |
| 9 | Streptococcus | 99.34 | 0.99 | 0.997 | 0.998 | 0.99 |
ProBioPred server is tested on browsers listed below:
| Operating System (Versions) | Chome (Versions) | Firefox (Versions) | Safari (Versions) | Microsoft Edge (Versions) |
|---|---|---|---|---|
| Linux (18.04 LTS) | 78.0.3904.108 (Official Build) (64-bit) | 70.0.1 (64-bit) | - | - |
| MacOS (10.13.4) | - | 70.0.1 (64-bit) | 11.1 | - |
| Windows (7, 10) | 78.0.3904.108 (Official Build) (64-bit) | 70.0.1 (64-bit) | - | 71.0.309.43 (64-bit) |
References:
- CChen L, Zheng D, Liu B, Yang J, Jin Q. VFDB 2016: hierarchical and refined dataset for big data analysis--10 years on. Nucleic Acids Res. 2016 Jan 4;44(D1):D694-7. doi: 10.1093/nar/gkv1239. Epub 2015 Nov 17.
- Jia B, Raphenya AR, Alcock B, Waglechner N, Guo P, Tsang KK, Lago BA, Dave BM, Pereira S, Sharma AN, Doshi S, Courtot M, Lo R, Williams LE, Frye JG, Elsayegh T, Sardar D, Westman EL, Pawlowski AC, Johnson TA, Brinkman FS, Wright GD, McArthur AG. CARD 2017: expansion and model-centric curation of the comprehensive antibiotic resistance database. Nucleic Acids Res. 2017 Jan 4;45(D1):D566-D573.doi: 10.1093/nar/gkw1004. Epub 2016 Oct 26.
- Lebeer S, Vanderleyden J, De Keersmaecker SC. Genes and molecules of lactobacilli supporting probiotic action. Microbiol Mol Biol Rev. 2008 Dec;72(4):728-64, Table of Contents. doi: 10.1128/MMBR.00017-08. Review.
- Chih-Chung Chang and Chih-Jen Lin, LIBSVM : a library for support vector machines. ACM Transactions on Intelligent Systems and Technology, 2:27:1--27:27, 2011 Software available at http://www.csie.ntu.edu.tw/~cjlin/libsvm
