Contents:
* ProBioPred Core Utility:
-
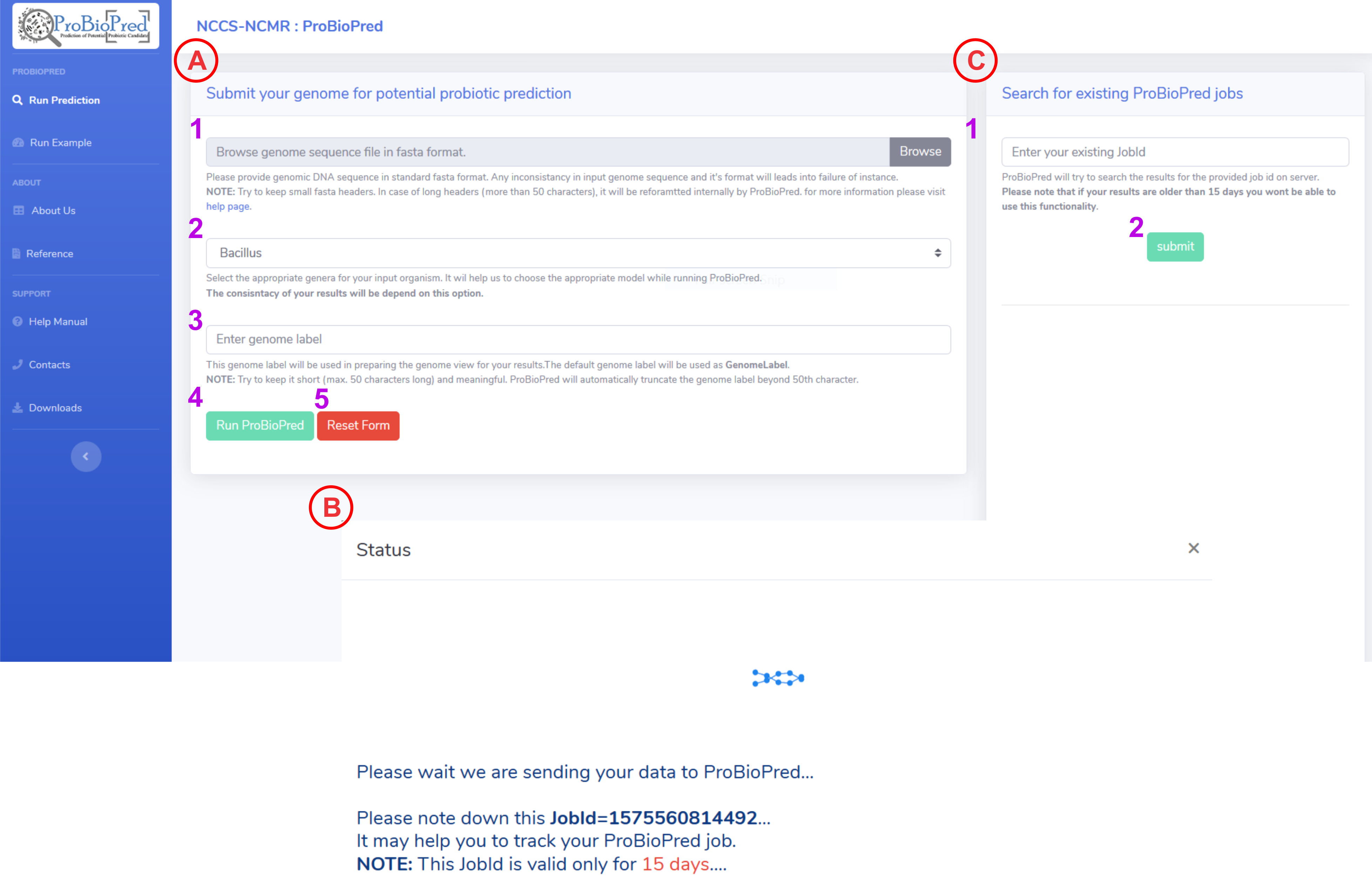
Submit your genome for the prediction:
-
To upload a genome of interest click on the Browse button and select a file from you local machine.
NOTE: Only nucleotide fasta will be taken as input - From the dropdown menu select an appropriate genus for the input genome (Bacillus, Bifidobacterium, Clostridium, Enterococcus, Lactobacillus, Lactococcus, Leuconostoc, Pediocoocus, Streptococcus). This option will help us to choose an SVM model for the prediction. Be cautious while selecting a genus as entire results are dependent on this choice.
-
Enter a genome label. This will be used in genome view for the results.
NOTE: Try to keep it short (max. 50 characters long) and meaningful. ProBioPred will automatically truncate the genome label beyond 50th character. - If you fill all the mandatory information then Run ProBioPred button will enable. After clicking this button form will be submit and your data will send to the server for processing.
- Reset the filled form by clicking rest button.
-
To upload a genome of interest click on the Browse button and select a file from you local machine.
-
Job Status:
-
After submitting the form, ProBioPred will generate an unique jobId. You can use this jobId to track your job.
NOTE: This jobId is valid only for 15 days after that the results will be removed from our server. -
Search Existing Jobs:
-
You can search for existing jobs within 15days time span using jobId.
- Enter an existing jobId (Ex: example)
- Click on search button.
* ProBioPred Log and Job Status:
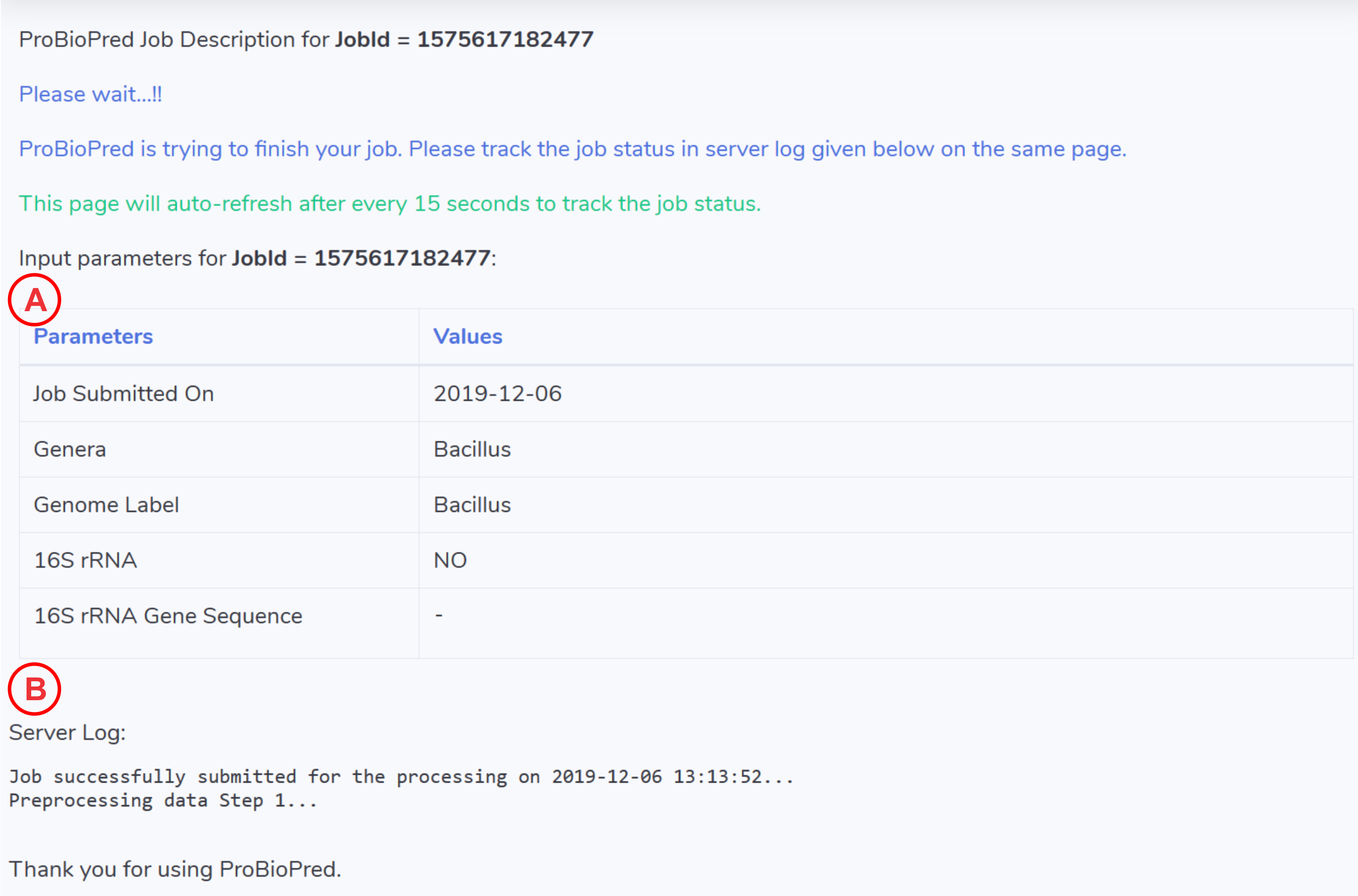
- Here you could find the input paramters used to run the job.
- This will be the live server log for the job submitted to ProBioPred server. The page will automatically refresh after every 15 seconds to update the log. You can keep track of your job here. If your job exited with error ProBioPred will report the erroneous step in the log.
* ProBioPred Results:
-
Result View Controller
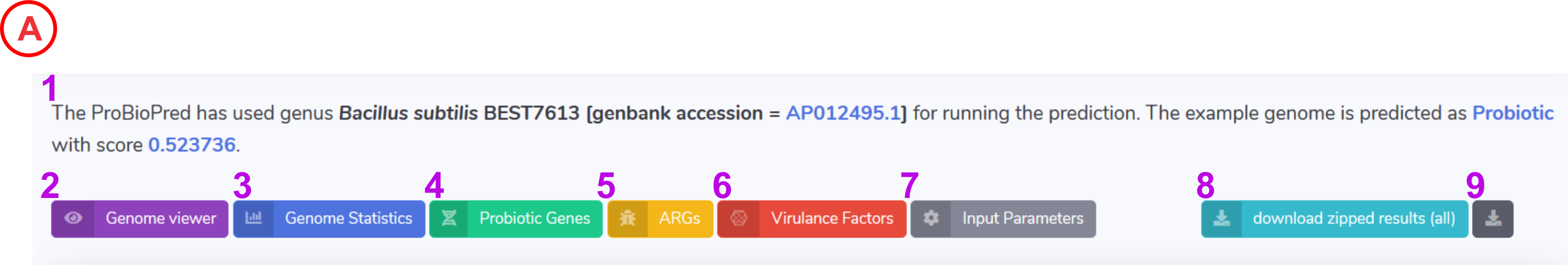
- The text will provide an overview of the ProBioPred result. It will report the ProBioPred score and the status of an input genome being probiotic or non-probiotic
- Clicking on this tab displays the Atlas genome view
- Clicking on this tab displays general genome statistics such as total number of sequences, N50, L50 etc.
- Clicking on this tab displays the probiotic gene annotation for the input genome.
- Clicking on this tab displays the available Antibiotic Resistance Genes (ARGs) present in the input genome.
- Clicking on this tab displays the identified Virulence Factors (VFs) present in the user supplied genome.
- Clicking on this tab displays the input parameters used to run the job.
- Clicking on this button will help to download the entire results in the .zip format.
-
Clicking on this button will help to download the complete active annotation table i.e. probiotic genes or ARGs or VFs or Input parameters in .xls format.
-
Genome View Atlas:
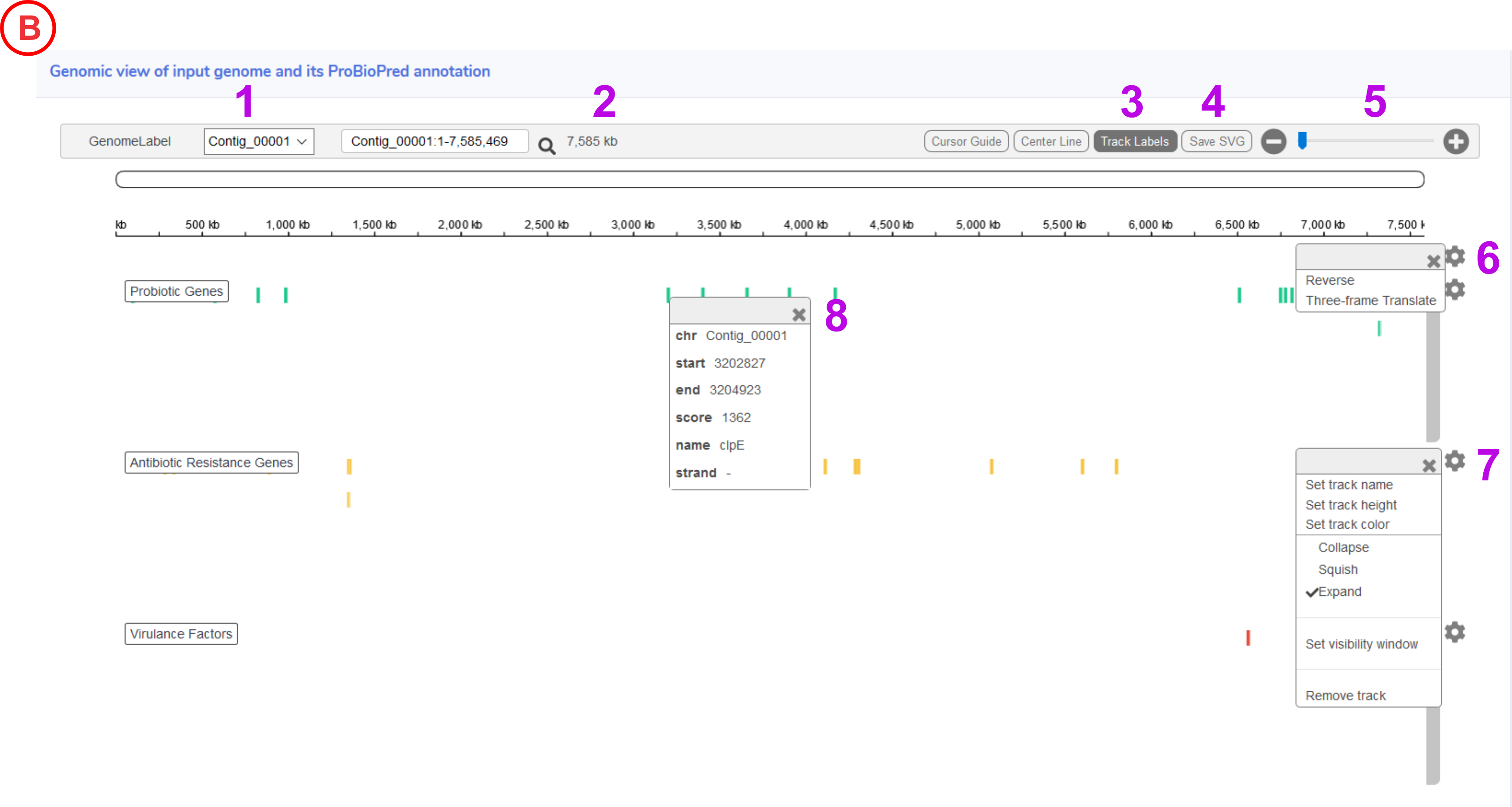
- Dropdown allow user to select the contig for visualization.
- Displays the size of active genomic region in the genome view.
- This button will help to hide and show the labels which are displayed on left side for the tracks i.e. Probiotic Genes, Antibiotic Resistance Genes and Virulence Factors.
- The entire displayed Genome Atlas image could be saved in .svg format.
- This Zoom bar, Zooms the genome view up to base pair view.
- This icon will help user to view the three frame translation and also the reverse view of the displayed contig.
- Clicking on the icon will help user to set change the track labels (Button 3) in various aspects such as colour height or even remove them.
- Clicking on the specific genomic region it will display the gene information such as location with start, end, gene name and strand.
-
Genome Statistics:
The tab Input Genome Statistics provides general genome information such as the total number of sequences, N50, % AT etc.
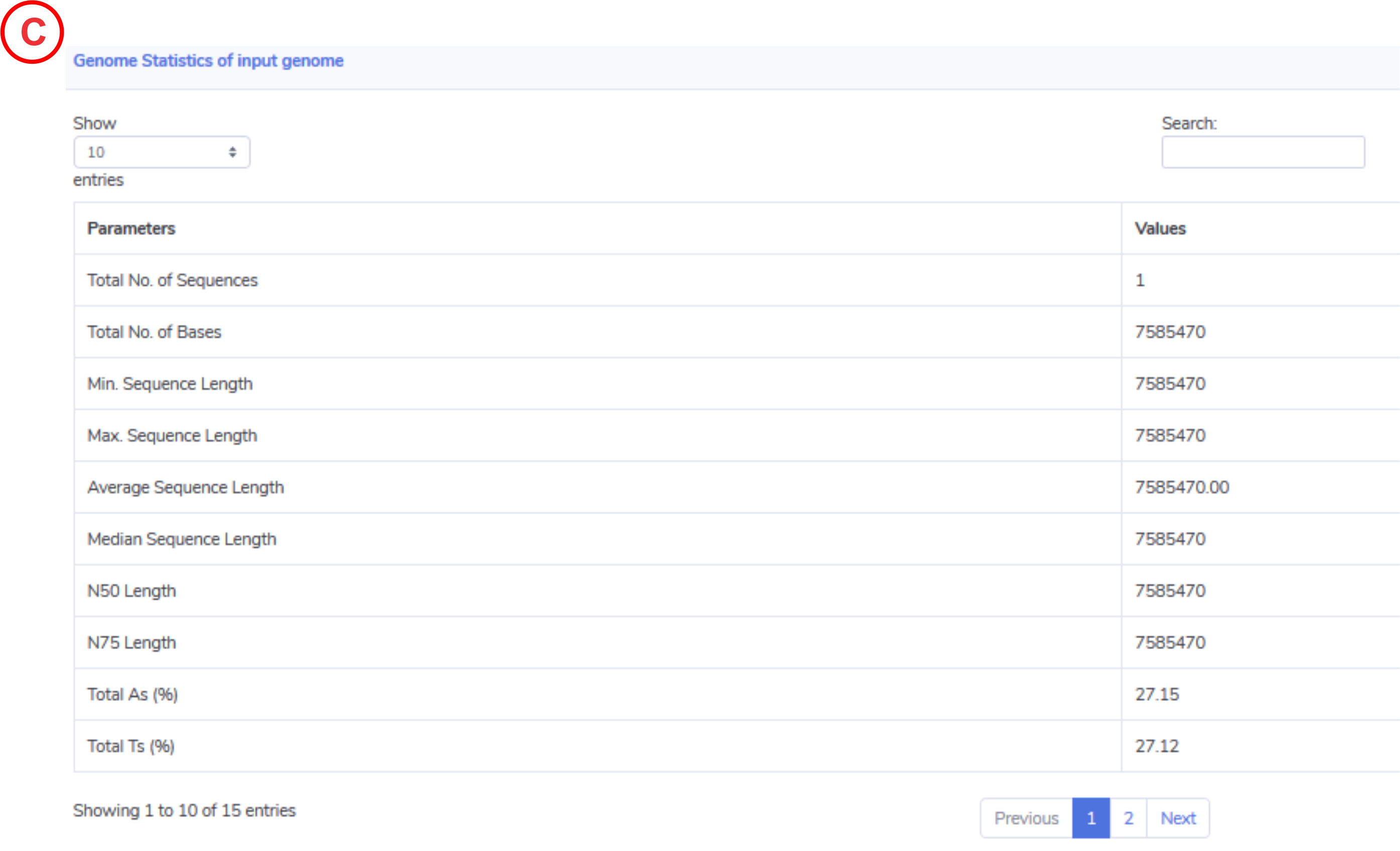
-
Probiotic Gene Content:
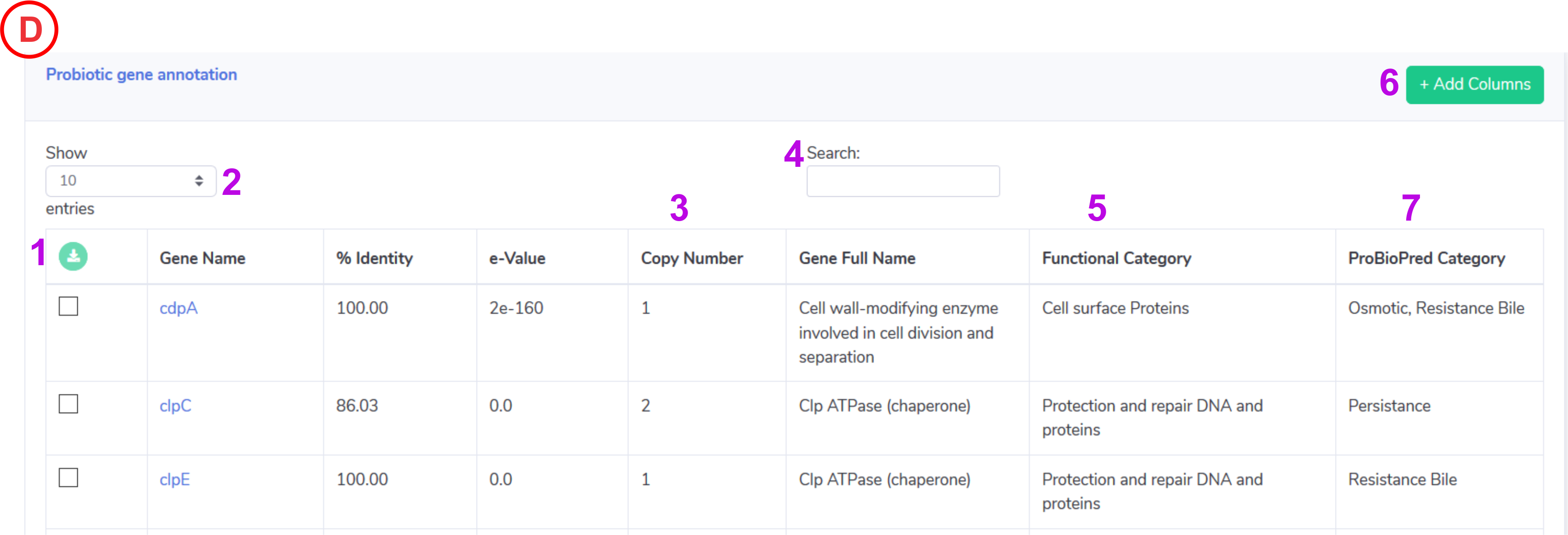
- Help to download the selected entire from the active annotation table in .xls format.
- Control the number of records to show in active annotation table.
- Detected copy number for the identified probiotic genes for the input genome.
- Help to search the keyword in active annotation table.
- Displays the gene’s Functional category.
- This will help to update the columns in active annotation table such as start, end , bit and query converge.
-
Displays the gene’s probiotic category (this could be the essential feature of an organism for being probiotic) assigned by ProBioPred.
NOTE: Displays the Gene name identified in the query genome, Clicking on the gene will take user to the UNIPROT page for detailed information
-
Antibiotic Resistance Gene Content:
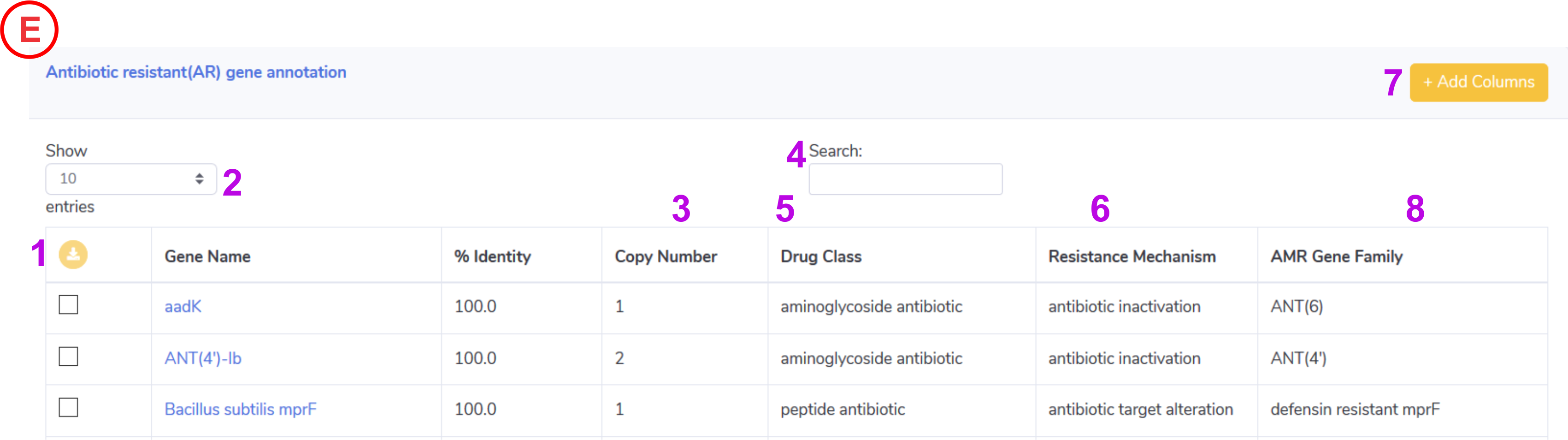
- Help to download the selected entire from the active annotation table in .xls format.
- Control the number of records to show in active annotation table.
- Detected copy number for the identified antibiotic resistant genes for the input genome.
- Help to search the keyword in active annotation table.
- Displays the Drug Class.
- Displays the Resistance Mechanism.
- This will help to update the columns in active annotation table such as start, end , bit and query converge.
-
Displays the AMR Gene Family.
NOTE: Clicking on the gene name will take user to the CARD database for further detailed information.
-
Virulence Factors:
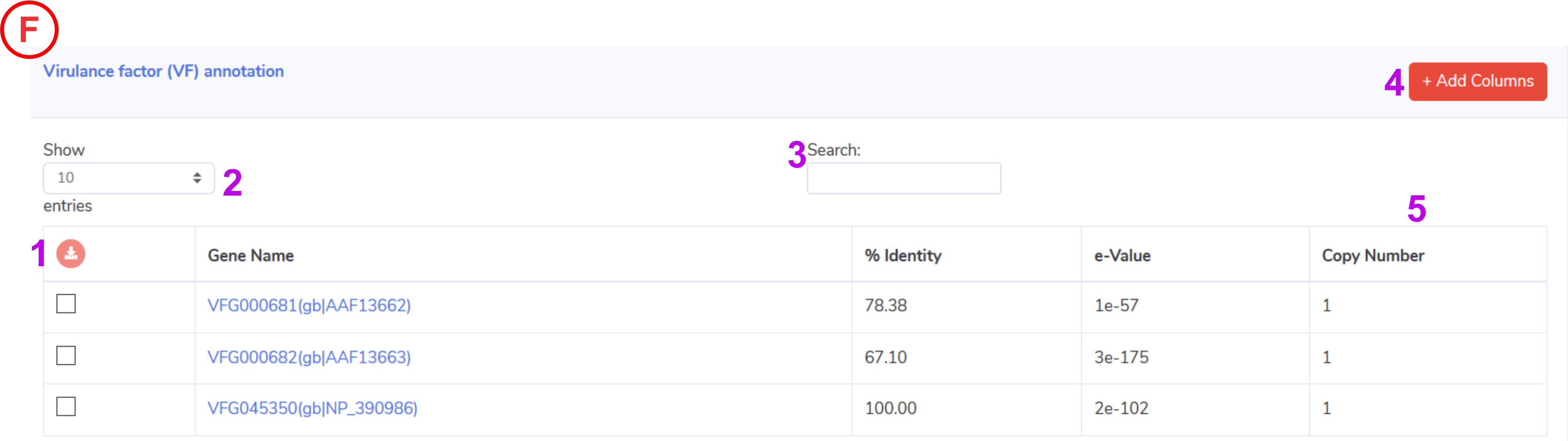
- Help to download the selected entire from the active annotation table in .xls format.
- Control the number of records to show in active annotation table.
- Help to search the keyword in active annotation table.
- This will help to update the columns in active annotation table such as start, end , bit and query converge.
-
Detected copy number for the identified virulance factors for the input genome.
NOTE: Clicking on the gene name will take user to the VFDB for further detailed information.
-
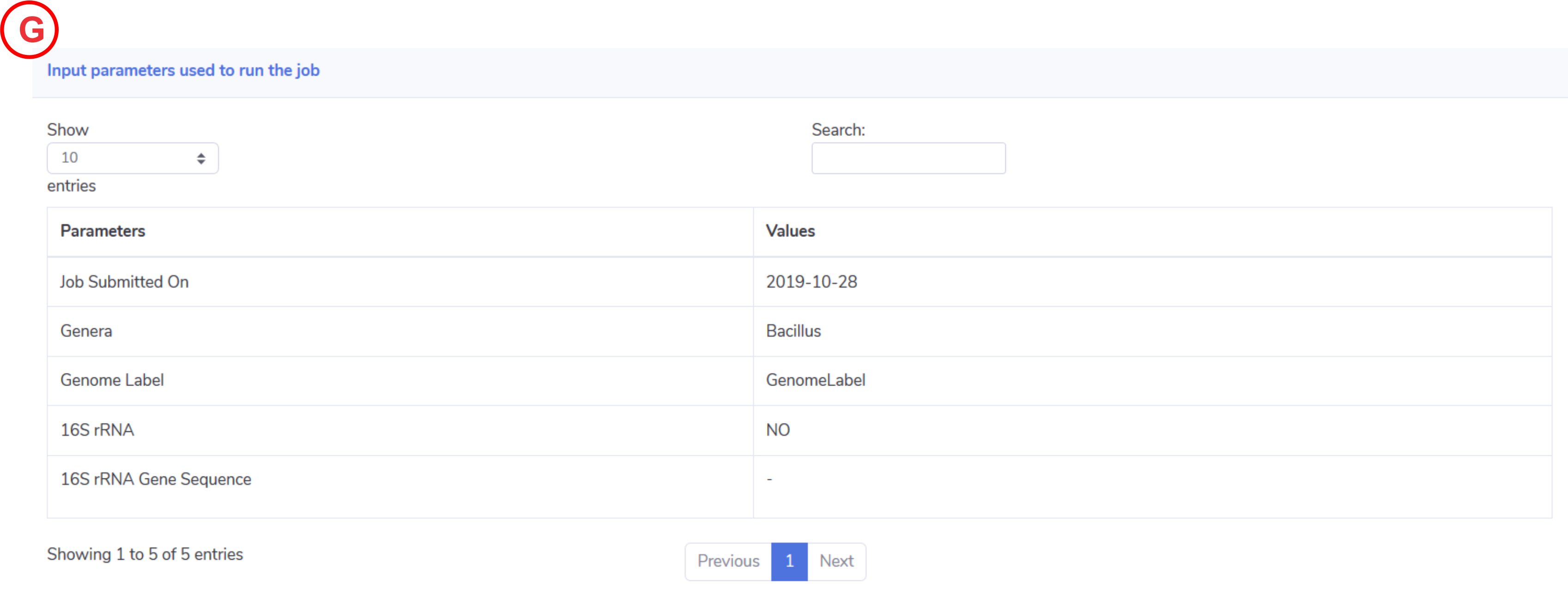
Input parameters:
This tab will display the parameters used to run the job.
* Process a genome which belongs to an unknown genera:
-
-
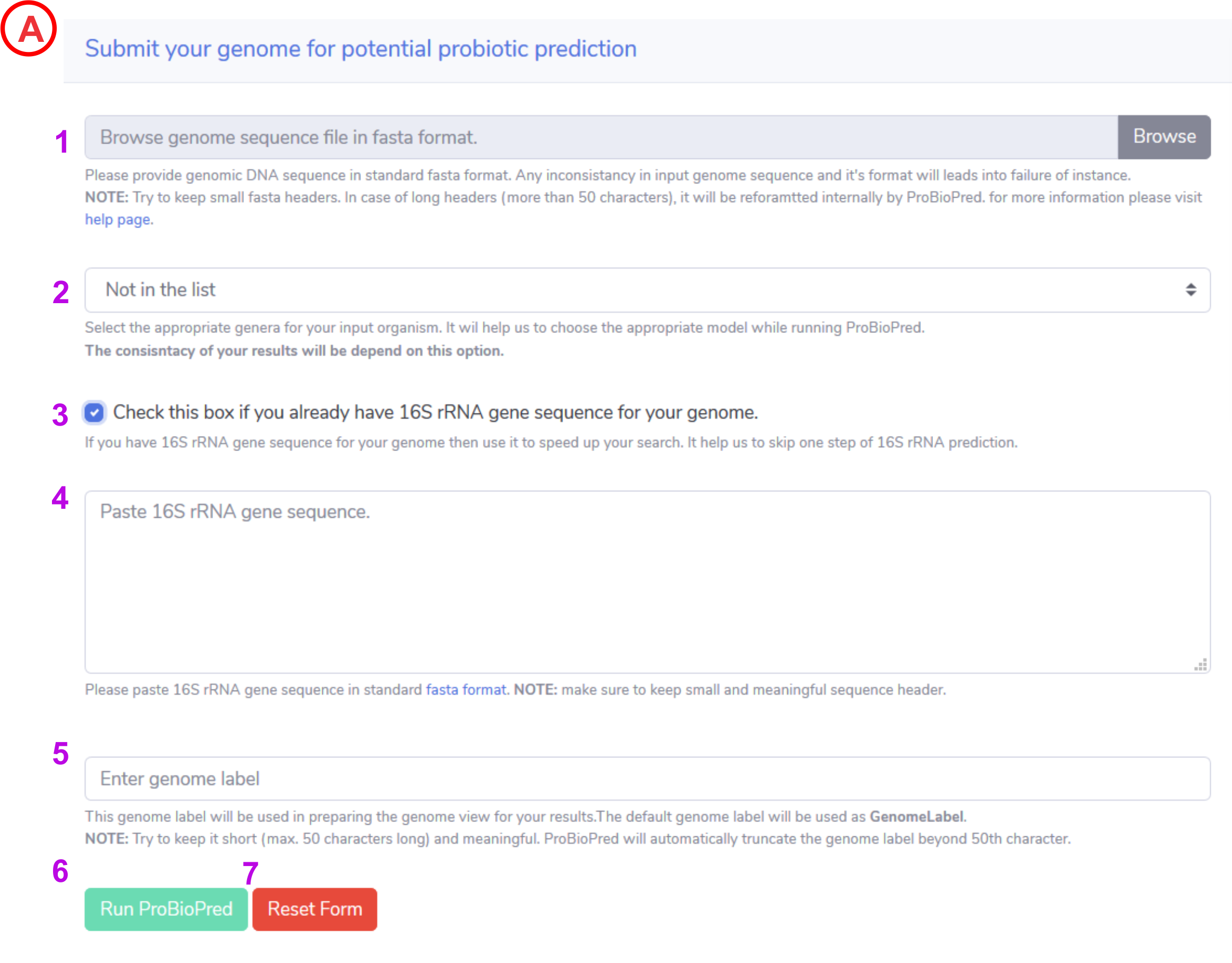
To upload a genome of interest click on the Browse button and select a file from you local machine.
NOTE: Only nucleotide fasta will be taken as input -
From the dropdown menu select an appropriate genus for the input genome (Bacillus, Bifidobacterium, Clostridium, Enterococcus, Lactobacillus, Lactococcus, Leuconostoc, Pediocoocus, Streptococcus). This option will help us to choose an SVM model for the prediction. Be cautious while selecting a genus as entire results are dependent on this choice.
If your uncertain about the genus under which the input genome belongs, you can select Not in the list option from dropdown. -
Not in the list option from above dropdown menu will view this check box. If you have 16S rRNA sequence for the input genome you can check this box otherwise ProBioPred will run rnammer and predict 16S rRNA sequence by it's own. This sequence will be used to determine the genus for input genome.
NOTE: We encourage users to provide an authentic 16S sequence for better results. -
If you have 16S rRNA gene sequence for your genome and if you checked the checkbox in previous step then this textbox will display. This will allow you to paste the 16S rRNA gene sequence.
NOTE: the entered 16S rRNA sequence should be in standard fasta format i.e. it should have valid header (first line starts with > sign) and subsequent lines shoulf be the nucleotide seqeuence. -
Enter a genome label. This will be used in genome view for the results.
NOTE: Try to keep it short (max. 50 characters long) and meaningful. ProBioPred will automatically truncate the genome label beyond 50th character. - If you fill all the mandatory information then Run ProBioPred button will enable. After clicking this button form will be submit and your data will send to the server for processing.
- Reset the filled form by clicking rest button.
-
To upload a genome of interest click on the Browse button and select a file from you local machine.
